The DNA [deoxyribonucleic acid]:
DNA is a long polymer of deoxyribonucleotides.
-
Bacteriophage φX174 has 5386 nucleotides.
-
Bacteriophage Lambda has 48502 base pairs (bp).
-
Escherichia coli has 4.6 x 106bp.
-
Haploid cell of human DNA contains 3.3x109 bp.
The structure of polynucleotide chain:
-
A nucleoside is the basic unit of polynucleotide chain of DNA or RNA.
-
Each nucleotide is composed of three components-a nitrogenous base,pentose sugar (ribose in case of RNA, and deoxyribose for DNA) and a phosphate group.
-
Nitrogenous bases are of two types- purine (adenine and guanine) and pyrimidines (Cytosine and thymine).
-
Uracil is only present in RNA instead of thymine.
-
A nitrogenous base is attached to the pentose sugar by an N- glycosidic linkage to form nucleoside.
-
When a phosphate group is attached to 5'-OH of a nucleoside through phosphodiester linkage, a nucleotide is formed.
-
Two nucleotides are joined through 3'-5' phosphodiester linkage and a dinucleotideis formed. When number of nucleotides are joined a polynucleotide chain is formed.
-
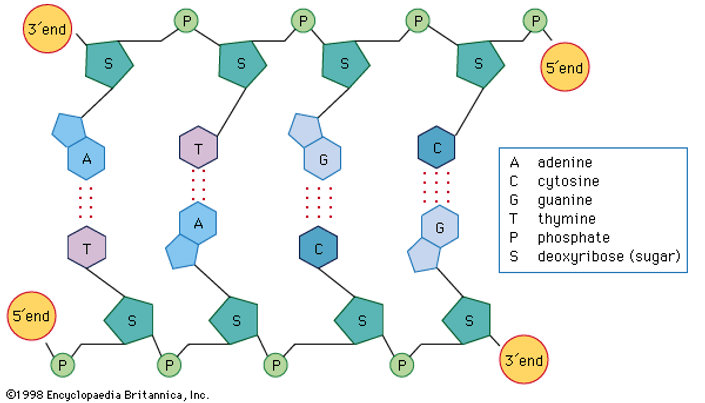
One end of the polynucleotide chain contains pentose sugar with the free OH at 5' (it is called 5'-end) and the other end contains sugar with free OH at 3' end (it is called 3'-end).
-
Sugar and phosphate constitute the backbone of polynucleotide chain and the nitrogenous bases are linked to sugar moiety which projects from the backbone towards inside.
The structure of double helical DNA:
The Double helix model of DNA was proposed by James Watson and Francis Crick in 1953 based on the X-ray diffraction data produced by Maurice Wilkins and the Rosalind Franklin and Erwin Chargaff's rules of base pairing.
Chargaff's rule:
-
The amount of adenine is always equal to the amount of thymine and the amount of guanine is always equal to the amount of cytosine, i.e., [A] = [T], [G] = [C].
-
Adenine is joined to thymine with 2 hydrogen bonds and guanine is joined two cytosine by 3 hydrogen bonds.
-
The ratio of adenine and guanine to that of thymine and cytosine is always equal to one.
Salient features of DNA:
-
DNA is made up of two polynucleotide chains, where the backbone is made up of sugar and phosphate groups and the nitrogenous bases project towards the centre.
-
The two Chains have anti-parallel polarity. It means, if one chain has the polarity 5'→3', the other has 3'→5'.
-
The bases in two strands are paired through hydrogen bond forming base pairs (bp). Adenine pairs with thymine by two hydrogen bonds and guanine pairs with cytosine with three hydrogen bonds. As a result, always a purine comes opposite to a pyrimidine.
-
The two chains are coiled in a right-handed fashion. The pitch of the Helix is 3.4 nm and there are roughly 10 bp in each turn.Consequently, the distance between a bp in a helix is approximately 0.34 nm.
-
The diameter of the strand is always constant due to pairing of purine and pyrimidine. This in addition to hydrogen bonds confers stability of the helical structure.
Watch Video
Central dogma:
Francis Crick proposed the central dogma in molecular biology, which states that the genetic information flows from DNA to mRNA (transcription) and then from mRNA to Protein (translation) always unidirectionally.
In some viruses the flow of information is in reverse direction, that is, from RNA to DNA. E.g., bacteriophage and HIV.
Packaging of DNA in prokaryotes:
-
In prokaryotes such as Escherichia coli, though they do not have a well-definednucleus , the negatively charged DNA is held with a positively charged non histone proteins to form nucleoid.
-
The DNA in nucleoid is organised in large loops held by proteins.
Packaging of DNA in eukaryotes:
-
In 1974, Roger Kornberg reported that chromosome is made up of DNA and protein. Later, Beadle and Tatum reported that chromatin fibres look like a beads-on-string, where beads or repeated unit of proteins.
-
There is a set of positively charged basic proteins called histones and acidic non-histone chromosomal [NHC] proteins.
-
Histones are rich in the basic amino acid residues lysine and arginine.
-
Histones are organised to form a unit of eight molecules called histone octomere.
-
The negatively charged DNA is wrapped around the positively charged histone octomere to form a structure called nucleosome.
-
A typical nucleosome contains 200 bp of DNA helix.
-
Nucleosome constitute the repeating unit of a structure in nucleus called chromatin, a thread-like stained bodies seen in nucleus.
-
The nucleosomes in chromatin are seen as‘beads-on-string’ structure under electron microscope.
-
The beads-on-string structure in chromatin is packaged to form chromatin fibres that are further coiled and condensed at metaphase stage of cell division to form chromosomes. The packaging of chromatin at a higher level requires additional set of proteins that collectively are referred to as non-histone chromosomal proteins [NHC].
Differences between euchromatin and heterochromatin

The search for genetic material:
Transforming principle:
-
In 1928, Frederick Griffith conducted a series of experiments with Streptococcus pneumoniae [a bacterium causing pneumonia].
-
He observed miraculous transformation in the bacteria during the course of his experiment, living organism (bacteria) had changed in physical form.
-
When bacterium grown on a culture plate, some produce smooth shiny colonies (S-strain) with mucous (polysaccharide) coat, while other produce rough colonies (R-strain) without coat or capsule.
-
When live S-strain bacteria were injected into mice, they died due to pneumonia.
-
When live R-strain bacteria injected into mice, they survived.
-
When heat killed S-strain bacteria were injected into mice, they survived and there were no symptoms of Pneumonia.
-
When he injected a mixture of heat killed S and live R-strain bacteria into mice, they died due to unexpected symptoms of Pneumonia and he recovered living as bacteria from the dead mice.
-
He concluded that R-strain bacteria had somehow been transformed by the heat killed S-strain bacteria. Some transforming principle, transferred from the heat killed S-strain, had enabled the R-strain to synthesise a smooth polysaccharide coat and become virulent. But he was not able to understand the cause of this bacterial transformation.
Biochemical characterization of transforming principle
-
Oswald Avery, Colin McLeod and Maclyn McCarty repeated Griffith's experiment in order to determine biochemical nature of transforming principle.
-
They concluded that DNA from the heat killed S-strain bacteria caused the transformation of non-violent R-strain bacteria into virulent S-strain bacteria.
-
They also discovered that proteases and RNases did not affect transformation while DNases Inhibited the process and they concluded that DNA is the hereditary material.

Proof for DNA as genetic material
-
Alfred Hershey and Martha Chase in 1952, conducted experiments on bacteriophage to prove that DNA is the genetic material.
-
Hershey and chase worked to discover whether it was a protein or DNA from the viruses that entered the bacteria, while they attach the bacteriophage to the bacteria.
-
They grew some viruses on a medium that contained radioactive phosphorus [32P] and some others on medium that contained radioactive sulphur [35S].
-
Viruses grown in the presence of radioactive phosphorus[32P] contained radioactive DNA.
-
Similarly, viruses grown in presence of radioactive Sulphur [35S] contained radioactive protein.
-
Both the Radioactive bacteriophages were allowed to infect E. coli separately.
-
Soon after infection, the bacterial cells were gently agitated in blender to remove viral coats from the bacteria.The virus particles were separated from the bacteria spinning them in a centrifuge.
-
By this experiment they concluded, bacteria which were infected with viruses that had radioactive DNA were Radioactive, indicating that that DNA was the material that passed from the virus to the bacteria.
-
Bacteria that were infected with viruses that had radioactive proteins were not Radioactive. This indicates that only DNA and not protein coat enter the bacterial cell. This proves that DNA is a genetic material which is passed from virus two bacteria and not protein.

Characteristics of genetic material
-
It should be able to generate its replica [replication].
-
It should be chemically and structurally stable.
-
It should be able to undergo slow and gradual change is called mutations which result in evolution.
-
It should be able to store genetic information which is inherited.
-
It should be able to express itself in the form of ‘Mendelian characters’.
Ribonucleic acid [RNA]
-
RNA was the first genetic material.
-
RNA behaved as a catalyst as well as genetic material.
-
RNA can synthesize DNA [reverse transcription] and it is estimated that DNA has evolved from RNA with the chemical modification.
-
The backbone of RNA is made up of a pentose ribose sugar and phosphate.
-
The 2'-OH group of ribose makes RNA labile and easily degradable.
-
RNA has two types of nitrogenous bases –
purines - adenine and guanine
pyrimidines - cytosine and uracil

DNA Replication
James Watson and Francis Crick in 1953 proposed scheme that DNA replication was semi- conservative.
The scheme suggested that the two strands would separate and act as a template for the synthesis of new complementary strands. After the completion of replication, each DNA molecule would have one parental and one newly synthesized strand. This scheme was termed as semi conservative DNA replication.
The experimental proof for semi-conservative mode of DNA replication
Matthew Meselson and Franklin Stahl in 1958 performed experiment on E. coli to prove the DNA replication is Semi-conservative.
-
They grew E. coli in a medium containing 15NH4Cl [in which 15N is the heavy isotope of Nitrogen] for many generations.
-
As a result,15N got incorporated into newly synthesized DNA.
-
This heavy DNA can be distinguished from the normal DNA by centrifugation in a Caesium chloride [CsCl] density gradient.
-
Then they transferred the cells into medium with normal 14NH4Cl and took the samples at various definite time intervals at the cells multiplied.
-
The extracted DNAs are centrifuged and measured to get their densities.
-
The DNA extracted from the culture after one generation from the 15N medium to 14N medium [E.coli divides every 20 minutes] showed an intermediate hybrid density.
-
The DNA extracted from the culture after two generations [that is after 40 minutes] showed equal amounts of light DNA and hybrid DNA.
-
Similar experiment was performed by Taylor and colleagues in 1958 on Viciafaba to prove the DNA in chromosome also replicate semi conservatively.

Enzymes for DNA replication
Various enzymes are required as catalysts during DNA replication in living cells.
-
DNA-dependent DNA-polymerase - It catalyzes the polymerization of deoxynucleotides on DNA template.
-
Helicase -It unwinds the DNA strand to form the replication fork.
-
DNA ligase -It joins the Okazaki fragments which are found on the lagging strand.
Mechanism or process of DNA replication
-
The unique and fixed point where the DNA replication originate is called ‘origin of replication’ or ‘ori’.
-
The complementary strands of DNA double helix are separated by DNA helicase and DNA gyrase. This process is called unwinding of double stranded DNA.
-
Unwinding of double stranded DNA forms a ‘Y-shaped’ configuration in the DNA duplex, which is called replication fork.
-
The DNA-dependent DNA-polymerase catalyse Polymerization only in one direction that is 5'→3'. This creates some additional complications at the replicating folk.
-
Consequently, on one strand [the template with polarity 3'→5'], replication is continuous, and it is called leading strand, while on the other [the template with polarity 5'→3'], it is discontinuous,and it is called lagging strand.
-
Replication of lagging strand generate small polynucleotide fragments called Okazaki fragments [after R. Okazaki, who first identify them].
-
These Okazaki fragments are then joined together by an enzyme called DNA ligase.

Transcription
-
The process of copying genetic information from one strand of the DNA into RNA is termed as transcription.
-
The principle of complementarity governs the process, except that adenosine now forms base pairs with uracil instead of thymine, as in replication.
-
Unlike replication, only a single stranded fragment of DNA gets copied into RNA.
Transcriptional unit
-
The transcriptional unit of DNA contains three regions in the DNA.A promoter, the structural gene and the terminator.
i. The promoter: it is the binding site for RNA polymerase for initiation of transcription.
ii. The structural gene: it codes for enzyme or protein for structural functions.
iii. The Terminator: it is the region where transcription ends.
-
DNA-dependent RNA-polymerase helps in DNA replication by catalysing the polymerization in only one direction i.e., 5'→3'.
-
The DNA strand that has the polarity 3'→5' acts as a template and is also referred to as template strand.
-
The strand which does not get transcripted is called coding strand and has the polarity 5'→3'. Its sequence is same as RNA formed.
-
The promoter is located towards 5' -end [upstream] of the structural gene of coding strands and provides the binding site for RNA polymerase.
-
The sequence of DNA located towards 3'-end [downstream] of the coding strand where the process of transcription would stop is called terminator.

Transcription unit and the gene
-
The segment of DNA coding for a polypeptide is called cistron.
-
In eukaryotes, the transcription unit possess a structural gene specific only for a single polypeptide. Thus, it is called monocistronic.
-
In prokaryotes, the transcription unit possessing the structural genes for many polypeptides which are part of single metabolic pathway are called polycistronic.
-
The gene in eukaryotes are split into the coding or expressed sequence of DNA called exon, and non-expressible sequence of DNA called intron.
-
mRNA contains only exon but no intron.
Transcription in prokaryotes
-
In prokaryotes, the structural gene is polycistronic and continuous.
-
In bacteria, the transcription of all the three types of RNA [mRNA, tRNA and rRNA] is catalysed by single DNA-dependent RNA-polymerase enzyme.
-
In E.coli bacterium, the RNA polymerase has co-factors α β along with the σ (sigma) factor, to catalyse the process.
-
The RNA polymerases able to catalyse all the three steps of transcription; they are, initiation elongation and termination.
-
Initiation: Sigma (σ) factor recognises the start signal and promoter region on DNA which then along with RNA polymerase binds to the promoter to initiate transcription.
-
Elongation:The RNA polymerase after initiation of RNA transcription loose the σ factor but continues the polymerization of ribonucleotides to form RNA.
-
Termination:Once the RNA polymerase reaches the termination region of DNA, the RNA polymerase is separated from DNA-RNA hybrid, as a result nascent RNA separate. This process is called termination which is the facilitated by a termination factor Rho (ρ).
-
In Prokaryotes, mRNA does not require any processing, so both the transcription and translation occur in the cytosol. It can be said that transcription and translation are coupled together.

Transcription in eukaryotes
-
In eukaryotes, the structural genes are monocistronic.
-
The process of transcription is similar to that in prokaryotes.
-
It takes place in the nucleus.
-
Coding gene sequences called exons from the part of mRNA and non-coding sequences called introns are removed during RNA splicing.
-
In eukaryotes, three types of RNA polymerase are found in the nucleus;
i. RNA polymerase I: transcribes rRNAs.
ii. RNA polymerase II: transcribes the precursor of mRNA [called heterogeneous nuclear RNA or hnRNA].
iii. RNA polymerase III: transcribes tRNA, 5SrRNA, and snRNA [ small nuclear RNAs].
Post transcriptional modifications
-
The primary transcripts are non-functional, containing both the coding regions, exon and non-coding region intron, in RNA and are called heterogenous RNA or hnRNA.
-
The hnRNA undergoes two additional process called capping and tailing.
-
In capping, an unusual nucleotide, methyl guanosine triphosphate (mGppp), is added to the 5'-end of hn RNA.
-
In tailing, adenylate residues (about 200 to 300) are added at 3'-end in a template independent manner.
-
Now the hnRNA undergoes a process where the introns are removed, and exons are joined to form mRNA by the process called RNA splicing.

Genetic code
-
The relationship between the sequence of nucleotides on mRNA and sequence of amino acids in the polypeptide is called genetic code.
-
George Gamow: suggested that the code must be of 3 bases in order to code for amino acids because there are only 4 bases (i.e., 43 or 4 X 4 = 64) which code for 20 amino acids.So, codon is a triplet.
-
Har Gobind Khorana:Developed chemical method for synthesizing RNA molecule with defined base combinations to develop the genetic code.
-
Marshall Nirenberg:Artificially synthesized proteins to understand the nature of codons.
-
Severo Ochoa:Demonstrated that polynucleotide phosphorylase also helped in polymerising RNA with defined sequences in a template-independent manner (enzymatic RNA synthesis).
Salient features of genetic code
-
The codons are triplet. Out of 64 codons, 61 code for 20 amino acids and 3 codons (UAA, UGA, UAG) do not code for any amino acid, hence function as stop or terminating codons.
-
One codon codes for only one particular amino acid, hence the code is unambiguous and specific.
-
Some amino acids are code by more than one codon, hence the code is degenerate.
-
The codon is read on mRNA in a continuous fashion, i.e., without punctuations and thus the code is comma less.
-
Genetic code is nearly universal,i.e., a particular codon codes for the same amino acid in all organisms except in mitochondria and few protozoa.
-
AUG is a dual function codon, it codes for methionine (met) and it is also acts as initiator codon.
Mutation
Mutation is defined as the sudden, stable, inheritable changes in the genetic materials of an organism.It can be of the following two major types;
-
Point mutation: It is the mutation in a single base pair, which is replaced by another base pair. For example, in Sickle-cell anaemia, point mutation in β-globin chain results in change of glutamate to valine.
-
Frameshift mutation: It is the change in the reading frame because of insertion or deletion of based pairs.
-
- It is the addition of one or more nucleotide in the DNA segment. Insertion of 3 or its multiple bases do not change the reading frame but add a new amino acid.
-
-It is the removal of one or more nucleotides from the DNA segment. Deletion of 3 or its multiple bases do not change the reading frame but remove one or more amino acids.
tRNA – The Adaptor Molecule
Francis Crick proposed the presence of an adaptor molecule which could read the codon one end and on the other end would bind the specific amino acids.
However, tRNA was known before genetic code was postulated and was then called sRNA (soluble RNA). Its role as an adapter molecule was reported later.
Structure of tRNA
-
The secondary structure of tRNA is clover-leaf like but the three-dimensional tertiary structure depicts it is a compact inverted ‘L’ shaped molecule.
-
tRNA has 5 arms or loops:
-
Anticodon loop: That has bases complementary to the code.
-
Amino acid acceptor end: To which amino acids bind.
-
T loop: Which helps in binding to ribosome.
-
D loop:Which helps in binding aminoacyl synthetase.
-
Variable loop
-
Each tRNA is specific for a particular amino acid.
-
A specific tRNA for initiation is called initiator tRNA.
-
There is no tRNA for stop codons.

Translation
-
Translation refers to the process of Polymerization of amino acids to form a polypeptide or it is the process of synthesis of protein from mRNA with the help of ribosome.
-
A translational unit in mRNA from 5'→3' comprises of a start codon, region coding for a polypeptide, a stop codon and untranslated region (UTRs) at both 5' -end and 3' -end for efficient process.
-
There are three stages of protein synthesis.
-
Initiation: Assembly of ribosome on mRNA. Activation of amino acids and its delivery to tRNA.
-
Elongation: Repeated cycle of amino acid delivery. Peptide bond formation and movement along the mRNA called translation.
-
Termination: The release of polypeptide chain.
Regulation of Gene Expression
-
Regulation of gene expression means controlling the amount and time of formation of gene products according to the requirements of the cell.
-
In eukaryotes, gene regulation can take place at four levels.
-
Transcription level (regulation of primary transcriptformation).
-
Processing level (regulation of splicing).
-
Transport of mRNA from nucleus to the cytoplasm.
-
Translation level.
The lac Operon
The concept of Operon was first proposed in 1961, by Jacob and Monod. An Operon is a unit of prokaryotic gene expression which includes coordinately regulated (structural) genes and control elements which are recognised by regulatory gene product.
Concept of an Operon
-
Structural gene: The fragment of DNA which transcribe mRNA for polypeptide synthesis.
-
Promoter: The sequence of DNA where RNA polymerase binds, and initiates transcription of structural genes is called promoter.
-
Operator: The sequence of DNA adjacent to the promoter where specific repressor protein binds is called operator.
-
Regulator genes: The gene that codes for the repressor protein that binds to the operator and suppresses its activity as a result of which transcription will be switched off.
-
Inducer: The substrate that prevents the repressor from binding to the operator, is called an inducer. As a result, transcription is switched on.It is a chemical of diverse nature like metabolite, hormone substrate etc...
The concept of lactose Operon (lac Operon)
-
It consists of regulatory genes, and structural genes lac ZYA.
-
Control genes consists of promoter. Between promoter and structural genes is the operator.
-
Regulatory gene lac I codes for repressor, which binds to operator and switches off oflac Operon.
-
When glucose not available, lactose available Lactose enters E.coli and binds inducer and forms inducer repressor complex that freese the operator.
-
RNA polymerase will transcribe the structural gene produce polycistronic mRNA now lac Operon is switched on.
-
Polycistronic mRNA is translated into three enzymes;
-
beta galactosidase-by Z,
-
betagalactosidepermease - by Y and
-
betagalactosidetransacetylase - by A

Human Genome Project [HGP]
-
The total cost of the project was 9 billion US dollar and an enormous amount of data was generated, therefore it is called a mega project.
-
The Human Genome Project was a 13-year project by the U.S. department of energy and the National Institute of Health. It was launched in 1990 and completed in 2003.
-
During the early years of Human Genome Project, the wellcome trust of UK became a major partner and additional contributions came from Japan,France, Germany and China.
-
HGP was closely associated with development of a new area in biology termed as bioinformatics.
-
If the obtained sequences HGP were to be stored in typed form in books, and if each page of the book contained 1000 letters and each book contained 1000 pages, then 3300 such books would be required to store the information of DNA sequence from a single human cell.
Goals of HGP
-
identify all the approximately 20,000 to 25,000 genes in human DNA.
-
Determine the sequences of the 3 billion chemical base pairs that make up human DNA.
-
Store this information in database.
-
Improvetools for data analysis.
-
Transfer related Technologies and information to other sectors, such as industries.
-
Address the ethical, legal and social issues [ELSI] that may arise from the project.
Methodologies of HGP
-
The methods involved two major approaches;
-
Expressed sequence tags (ESTs): This method focuses on identifying all the genes that are expressed as RNA.
-
Sequence annotation: It is an approach of simply sequencing the whole set of genomes that contains all the coding and non-coding sequences, and later assigning different regions in the sequence with functions.
-
For Sequencing, the total DNA from cell is first isolated and broken down in relatively small sizes as fragments.
-
These DNA fragments are cloned in suitable host using suitable vectors. When Bacteria is used as vector, they are called bacterial artificial chromosome (BAC) and when yeast is used as vector, they are called yeast artificial chromosomes (YAC).
-
Frederick Sanger developed a principal according to which the fragments of DNA Sequenced by automated DNA sequences.
-
On the basis of overlapping regions on DNA fragments, these sequences are arranged accordingly.
-
For alignment of these sequences, specialized computer-based programs were developed.
-
Finally, the genetic and physical maps of the genome were constructed by controlling information about the certain repetitive DNA sequences and DNA polymorphism, based on endonuclease recognition sites.
Salient features of Human Genome
-
The Human Genome contains 3164.7 million bases pairs.
-
The average gene consists of 3000 bases; the largest known human gene being dystrophin at 2.4 million bases.
-
The total number of genes is estimated to be 30,000 and 99.9% nucleotide bases are exactly the same in all people.
-
The functions are unknown for over 50% of the discovered genes.
-
Less than 2% of the genome codes for proteins.
-
The Human genome contains large repeated sequences.
-
The repeated sequence is thought to have no direct coding functions, but they throw light on chromosome structures, dynamics and evolution.
-
Chromosome 1 has most genes (2968) and the Y has the fewest genes (231).
-
Scientists have identified about 1.4 million locations where single base DNA sequence differences called single nucleotide polymorphism or SNPs occur in humans
Applications and future challenges
-
Complete the sequence of Human Genome when properly studied, will enable to develop radically new approach to biological research.
-
All genes in a genome and transcripts in a particular tissue, organ or tumor can be studied.
-
It enables us to understand how enormous number of genes and proteins work together.
DNA fingerprinting
-
DNA fingerprinting uses short nucleotide repeat called variable number tandem repeats (VNTRs) as Markers.
-
VNTRs vary from person to person and are inherited from one generation to the next. Only closely related individuals have similar VNTRs.
-
Dr. Alec Jeffreys developed the technique of DNA fingerprinting in an attempt identify DNA markers for inherited diseases.
Methodology and technique
-
DNA is isolated and extracted from the cell or tissue by centrifugation.
-
By the process of polymerase chain reaction (PCR), many copies are produced. This step is called amplification.
-
DNA is cut into small fragments by treating with restriction endonucleases.
-
DNA fragments are separated by agarose gel electrophoresis.
-
The separated DNA fragments are visualised under ultraviolet radiation after applying suitable dye.
-
The DNA is transferred from electrophoresis plate to nitrocellulose or nylon membrane sheet. This is called Southern blotting.
-
VNTR probes are now added which bind to specific nucleotide sequences that are complementary to them. This is called hybridisation.
-
The hybridised DNA fragments are detected by autoradiography. They are observed as dark bands on X-Ray film.
Applications of DNA fingerprinting
-
It is used as a tool inference tests to identify criminals and criminal investigations.
-
It is used to settle paternity disputes and maternity disputes.
-
It is used to determine population and genetic diversities to study evolution.
-
It is used in the study of evolutionary biology.

